Parallel-Meta Suite
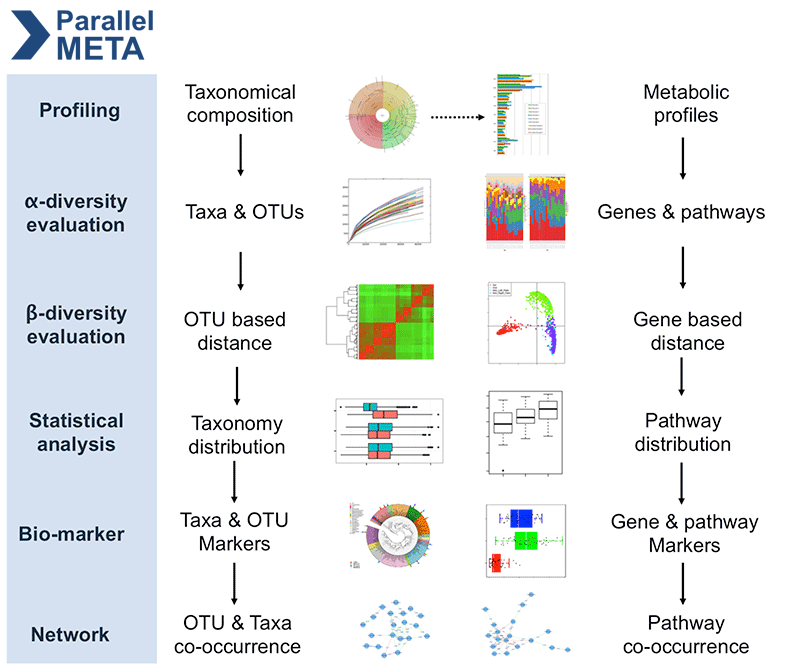
Parallel-Meta Suite is a comprehensive and full-automatic computational toolkit for rapid data mining among metagenomic datasets. Both metagenomic shotgun sequences and 16S/18S/ITS rRNA amplicon sequences are accepted. Based on parallel algorithms and optimizations, Parallel-Meta Suite can achieve a very high speed compare to traditional microbiome analysis pipelines. Now the Parallel-Meta Suite version 3.7.2 is available at http://bioinfo.single-cell.cn/parallel-meta.html.
Download The following functions have been updated in the version 3.7.2 of Parallel-Meta Suite.
- Add new databases, including GreenGenes 2 and RefSeq
- Add PM-profiler as a new taxonomy profiler
We strongly recommend all users to install/update the latest version.
3.7.3 (July 3, 2024)
Linux X86-64 (src package) Mac (Intel/M2/M3) (src package) You can also get the lastest release from Parallel-Meta Suite GitHub respository.
- System requirement
- Mac OS X needs to install the compiler that supports OpenMP by
brew install gcc- Quick installation
- Type the following commands in the directory that contains the package for quick installation:
tar -xzvf parallel-meta-3.7-src.tar.gz#Extract the package cd parallel-meta#Go to the package directory source install.sh#Automatic installation - The example dataset could be found at “example” folder. Check the “example/Readme” for details about the demo run.
Tutorial and Demo dataset Here we provide a demo dataset with 20 environment microbiome samples in five different environment(feces, tongue, palm, soil, marine). See the tutorial and"Readme" in the dataset package for details.
Sample Dataset and Sample output (by 3.7.2, GreenGenes 13-8 97% level, Vsearch)
Publications 1. Chen., et al., Parallel‐Meta Suite: Interactive and rapid microbiome data analysis on multiple platforms, iMeta, 2022.
2. Jing., et al., Parallel-META 3: Comprehensive taxonomical and functional analysis platform for efficient comparison of microbial communities, Scientific Reports, 2017.
3. Su X., Pan W., et al.,Parallel-META 2.0: Enhanced Metagenomic Data Analysis with Functional Annotation, High Performance Computing and Advanced Visualization, PLoS One, 2014.
4. Su X., et al. Parallel-META: Efficient Metagenomic Data Analysis Based on High-Performance Computation, BMC Systems Biology, 2012.
