Parallel-QC
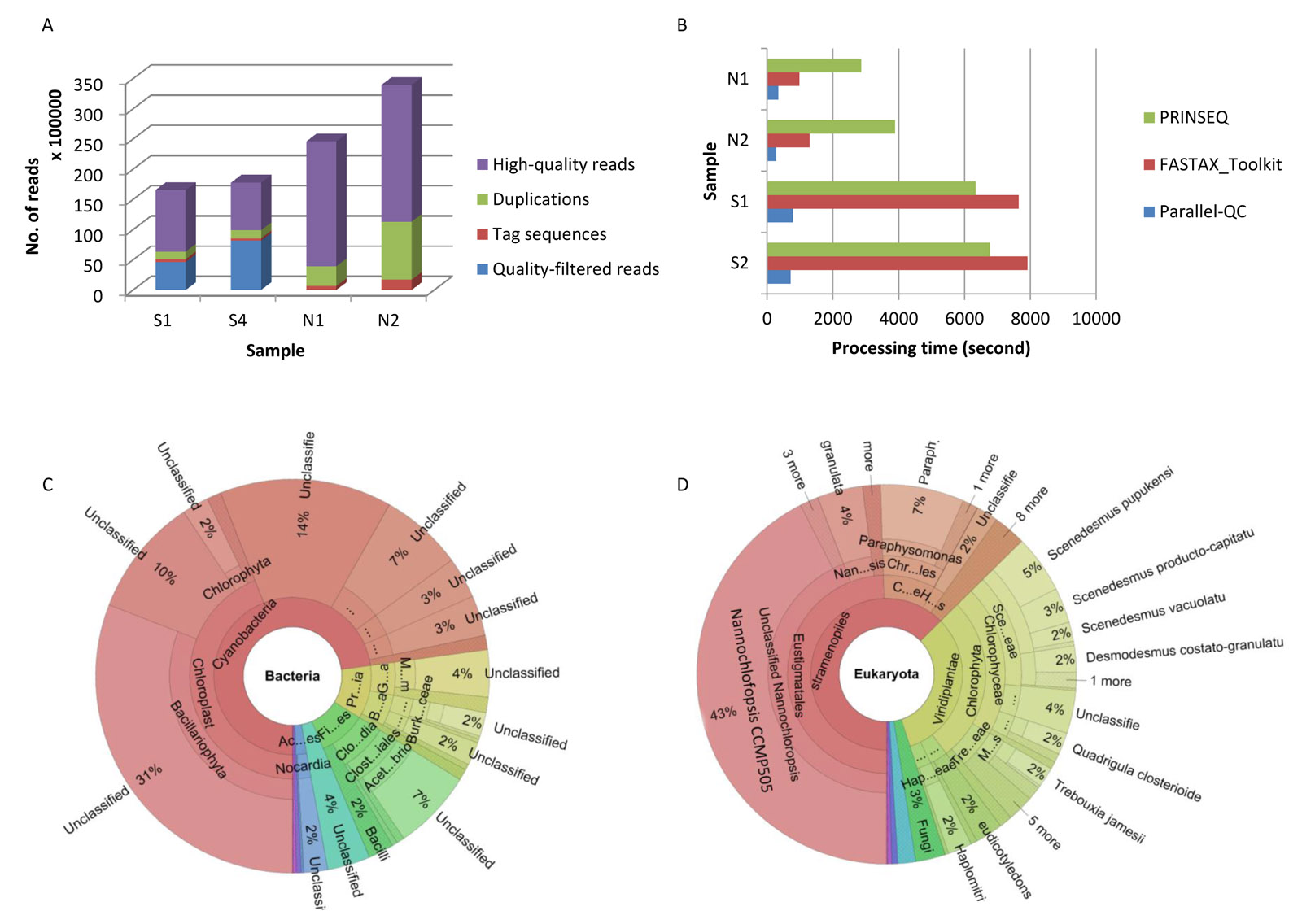
Quality control (QC) is the critical first step for processing raw next-generation sequencing (NGS) data, in which sequencing artifacts, including low-quality reads and contaminating reads, would significantly affect and mislead the results of downstream analysis. Here we report Parallel-QC, a fast computational engine specifically designed for general NGS data QC. Parallel-QC can complete sequencing-quality assessment accuracy and efficiency. Possible contaminating species could also be identified without any prior information. And the whole processing of Parallel-QC is quite fast since it is optimized based on parallel computation.
Parallel-QC Version 1.0 (Users' manual included)
Parallel-QC Development Kit Version 1.0 (Development manual included)