QIBEBT Proposed High-Performance Computational Method to Significantly Accelerate Large-scale Microbial Community Analysis
Metagenomics method directly sequences and dynamically analyses taxonomic and functional information of microbial communities, which could potentially optimize bio-processes behind some key bioenergy applications such as cellulose degradation, polysaccharide metabolism and biogas production. Data-mining among microbial community samples from different time and environment can discover valuable biological information hidden in the massive metagenomic data. However, current low-throughput metagenomic data analysis process has hampered large-scale microbial community analysis, thus it becomes the bottle neck in current metagenomic research.
Assistant professor SU Xiaoquan and WANG Xuetao, from Bioinformatics Group of Single Cell Center, at Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT), have proposed highly efficient algorithm name GPU-Meta-Storms to evaluate the similarities among microbial communities based on General-Purpose computing on Graphics Processing Units (GPGPU). This method provides a parallel computing solution that can compare a pair of microbial community samples within one micro second, which enables real-time analysis and monitoring for microbial communities. This work has been published in Bioinformatics online in Dec. 2013. In addition, the Bioinformatics Group has also obtained 5 software authorities on relative works.
This work is led by Prof. NING Kang of QIBEBT, and partially supported by the QIBEBT-NVIDIA Compute Unified Device Architecture (CUDA) Research Center.
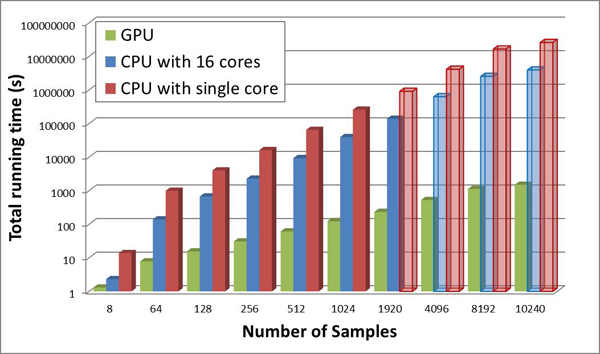
Figure 1. GPU-Meta-Storms provides a parallel computing solution for the high efficient analysis among massive number of microbial community samples, which could thus enable real-time monitoring of microbial communities. (Image by Single Cell Center at QIBEBT)
Reference:
Contact:
Prof. NING Kang
Email: ningkang (AT) qibebt.ac.cn
